
 

|

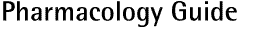
Molecular Pharmacology: Definitions
3 prime end (3')
'Back end' of the DNA or RNA strand. It refers to the direction of transcription or
translation (fig. H).
5 prime end (5')
'Front end' of the DNA or RNA strand. It refers to the direction of transcription or
translation (see fig. H).
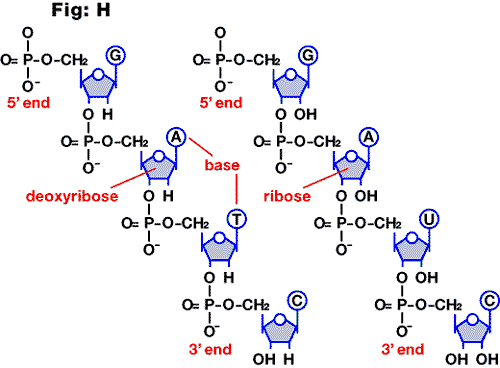
Fig. H: Structure and orientation of RNA and DNA
Alternative RNA splicing
Removal of all the introns and one or more exons in the RNA strand; can vary from cell
to cell.
Annealing
Base pairing of complementary single strands of DNA or RNA into a helix.
Anticoding DNA strand
Template that directs the synthesis of mRNA which is complementary to it. Synonymous
with antisense strand.
Anticodon
Complementary sequence on tRNA to an amino acid codon sequence.
Antisense strand
See anticoding DNA strand.
Base pair
This is the partnership of nucleotides within DNA or between DNA (A-T, C-G; see fig.
I) and RNA (A-U, C-G). Also used to describe the size of RNA or DNA or probe. Commonly
abbreviated to bp. Larger pairings are referred to as kilobases (Kb).
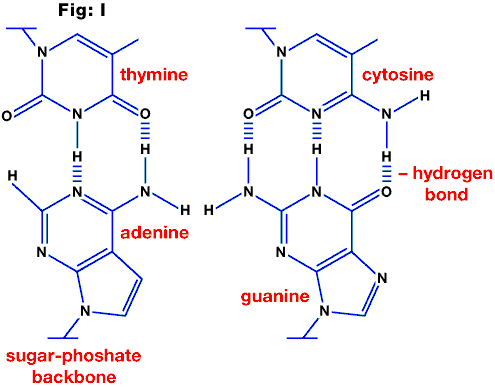
Fig. I
Base-pairing in DNA by hydrogen bonding.
cDNA
Complementary DNA. DNA synthesized from a RNA template by reverse transcriptase to
form an antisense 'copy' of the RNA.
cDNA library
Contains all the cDNA from a given organism, tissue or cell.
Central dogma
The usual direction of transfer of information from DNA to RNA to protein (see fig. J).
Clone
A collection of genetically-identical DNA fragments, cells or whole organisms.
Fig. J: Transfer of genetic information in eukaryotic cells.
Cloning vector
A plasmid or phage used to 'carry' and introduce foreign DNA into bacteria or
mammalian cells for the purpose of production of a specific protein.
Coding DNA strand
Contains the same sequence as mRNA; synonymous with sense
strand.
Codon
A triplet of nucleotides representing an amino acid or termination signal.
Cosmid vector
Plasmid vector into which cos sites have been inserted
which can incorporate long sequences of foreign DNA.
Cos site
Acronym from COhesive end Site. A region of DNA which allows large pieces of
chromosomal DNA to be incorporated into a plasmid vector (known in this instance as a cosmid vector) during DNA replication.
Denaturation
Used with reference to double-stranded DNA to mean the separation of the two
complementary strands.
Domain
A section of a gene pertaining to a specific amino acid sequence or function.
DNA
Deoxyribonucleic acid (see fig. H).
DNase
Enzyme that digests DNA.
DNase 1
A nuclease which cleaves specific nucleotides in a piece of DNA. It is only found
within the nucleus.
DNA ligase
Joins together (ligates) two double stranded DNA fragments.
DNA polymerase 1
Enzyme used to synthesize new DNA strands from a template.
Dot blotting
Technique used to identify DNA which has been spotted (dotted) onto filter paper.
Hybridization of this DNA is then achieved with radiolabelled DNA or RNA probes.
Endonuclease
An enzyme that cleaves bonds within a nucleic acid chain.
Exon
Nucleotide segment within the DNA sequence of a gene encoding for amino acids (see fig. J).
Exonuclease
An enzyme that cleaves whole nucleotides from either the 5' or 3' end of the RNA or
DNA chain.
Expression
The transcription and translation of a genetic message to protein.
Expression Vector
A cloning vector 'designed' to allow a particular DNA coding sequence to be
transcribed and thus translated into protein.
Genome
Entire genetic DNA material from a single organism.
Genomic library
A set of cloned fragments from a single organism's genome.
Hybridization
Pairing of complementary RNA and/or DNA.
Inducer
Agent that induces gene transcription via interaction with a regulating protein.
In situ hybridization
Hybridization performed on sections of tissue/cells mounted on slides using labelled
DNA or RNA probes.
Intron
Intervening sequence between exons in DNA which does NOT code for protein (see fig. J).
Library
Set of cloned fragments which, together, represent an entire genome.
Ligation
The joining together of two double-stranded molecules of DNA by DNA ligase.
mRNA
Messenger ribonucleic acid. This is a single stranded base chain which is responsible
for transferring coded information from the nucleus to the cytoplasm to specify the
production of proteins. Comprises 2% of total cell RNA.
Nick translation
A process for radiolabelling DNA in which the starting point, on a DNA strand, is
identified ('nicked') for degradation, and then subsequently replaced with radiolabelled
nucleotides. This process is catalaysed by DNA polymerase 1 and DNase.
Nonsense codons
Serve as chain terminating 'stop' signals for protein synthesis.
Northern Blot
Size separation of RNA by electrophoresis followed by
transfer to nitrocellulose filter paper or membranes and subsequent detection of a
specific RNA sequence by a cDNA probe. Used to identify separated RNA with complementary
pieces of DNA.
Nuclease S1
An enzyme which degrades single-stranded DNA regions.
Nucleotide
One of 5 bases which form DNA (ACGT) or RNA (ACGU):- adenosine (A), cytosine (C),
guanine (G), thymidine (T) or uridine (U).
Oligonucleotide
(commonly abbreviated to 'oligo'). A short RNA or DNA strand up to 100 bases in
length. Used as a probe in hybridization studies.
Oncogene
Normal cellular genes which, when inappropriately expressed or mutated, can transform
eukaryotic cells into tumour-like cells.
Operon
A cluster of functionally-related genes, regulated and transcribed as a unit.
PCR
Polymerase chain reaction. A method for detection of nucleic acids by greatly
amplifying the number of specific sequences of DNA.
Phage
A bacterial virus.
Plasmid
A self-replicating piece of circular DNA.
Poly-A tail
This is the multiple addition of adenosine (A) groups on the 3' end of a piece of RNA,
after transcription.
Primer
A short nuclei acid fragment used fro the initiation of DNA synthesis with DNA
polymerase.
Probe
A labelled (radioactive) piece of single stranded DNA/mRNA which can be used to
locate/identify (i.e. hybridize with) its complementary mRNA/DNA.
Promoter region
Sequence of DNA that is responsible initiating RNA synthesis by binding RNA
polymerase.
Proto-oncogene
Abbreviated commonly to c-onc (cellular) or v-onc (viral). This is the normal
counterpart in the genome to oncogenes found in some retroviruses.
Recombinant DNA
DNA which has been artificially combined from separate segments.
Repressor
A molecule which binds to DNA and prevents transcription of RNA and hence synthesis of
a particular protein.
Restriction enzyme
Cuts DNA at specific sequences to produce varying lengths of the original base chain.
Restriction map
Map showing points where restriction enzymes can cut DNA strand.
Retrovirus
RNA virus that replicates itself via conversion into duplex DNA.
Reverse transcriptase
Enzyme that catalyses the synthesis of cDNA from a RNA template.
RNA
Ribonucleic acid (see fig. H).
RNA polymerase
Enzyme involved in the transcription of DNA into its complementary mRNA.
RNase
Enzyme that digests RNA.
RNA splicing
The step in RNA maturation in which introns and removed and exons are linked together.
rRNA
Ribosomal RNA associated with protein production within the ribosomes. Comprises 93%
of total cell RNA.
Sense strand
Sequence of DNA with an identical sequence to RNA.
Southern Blot
Used to identify DNA which has been digested with restriction enzymes and separated
onto a gel and transferred for hybridization with complementary pieces of RNA.
Split gene
A gene containing introns.
Transcription
Synthesis by RNA polymerase of mRNA from DNA. It occurs from the 5' to the 3' end.
Transduction
The transfer of genes from one bacterium to another by a bacteriophage.
Transfection
The introduction of bacteriophage DNA into a second vector.
Translation
Synthesis of polypeptides by ribosomes from genetic information carried by mRNA.
tRNA
Transfer RNA. Involved in bringing amino acids to the ribosome for protein
construction. Constitutes 5% of total cell RNA.
Vector
See cloning vector.
Western Blot
Used to identify proteins which have been separated on a gel by using specific
antibodies
 Definition index Definition index
 Table of Contents Table of Contents
|



![]() Definition index
Definition index![]() Table of Contents
Table of Contents